VKORC1 and CYP2C9 Genotype Variations in Relation to Warfarin Dosing in Korean Stroke Patients
Article information
Abstract
Background and Purpose
Variant alleles of CYP2C9 and VKORC1 account for differences in anticoagulation response. We sought to establish a warfarin dosing formula for individualized target International Normalization Ratio of Prothrombin Times (INRs) using data from single nucleotide polymorphisms (SNPs) in VKORC1 and CYP2C9 in Korean patients.
Methods
Ischemic stroke patients displaying stable target INR for at least 3 months before enrollment were analyzed. Warfarin and vitamin K levels were measured to adjust for confounders. Phenotypes were defined using the 'warfarin response index' (WRI) defined as INR divided by the daily maintenance warfarin dose. We tested SNPs in CYP2C9 (3 sites: 430C>T (rs1799853), 1075A>C (rs1057910), 1076T>C) and VKORC1 (14 sites: 381C>T, 861C>A (rs17880887), 2653G>C, 3673A>G, 5496G>T, 5808T>G (r17882154), 6009C>T, 6484T>C (rs9934438), 6853C>T (rs17886369), 7566T>C, 8767G>C, 8814T>C, 9041G>A (rs17880624), and 9071G>T) using a standard sequencing method. Multivariate linear regression analysis was applied to establish the formula for warfarin dosage.
Results
All 204 patients had excellent drug compliance. The mean INR was 2.22 (+0.56) and mean daily maintenance dose of warfarin was 3.92 mg (+1.54). Patients with low WRI were younger (P<0.001) with high body mass index (P=0.003), high prevalence of wild-type CYP2C9 polymorphism (1075A>C, P<0.001), and six heterozygote SNPs in VRORC1 (P<0.001), which were tightly interlinked (381T>C, 3673G>A, 6484T>C, 6853C>G. 7566C>T, 9041G>A) (r2=1). Based on these data, a warfarin dosing formula was established.
Conclusions
WRI is influenced by age, body mass index and SNPs in VKORC1 and CYP2C9 in Korean stroke patients. The obtained warfarin dosing formula may be clinically applicable.
Introduction
Warfarin is the most widely used anticoagulant in stroke patients. Oral anticoagulation is superior to antiplatelet therapy involving clopidogrel or aspirin in the secondary prevention of stroke in patients with cardiogenic embolism.1,2 However, warfarin has a narrow therapeutic index, particularly for stroke patients; a suboptimal International Normalization Ratio of Prothrombin Time (INR) level would increase stroke recurrence while excessive anticoagulation results in brain hemorrhage. Considering the high mortality of warfarin-induced brain hemorrhages, proper adjustment of anticoagulation intensity is critical for the effective management of stroke patients.
Age, gender, concurrent medications, and dietary vitamin K intake influence the INR level,3-7 and algorithms using these parameters have been developed.6,8 Nevertheless, these factors are insufficient to explain the wide inter-individual differences in doses required for optimal INR,9-11 which may be related to individual genetic variability.11-14 More recent studies have demonstrated that variant alleles of Vitamin K expoxide reductase complex, subunit 1 (VKORC1) and Cytochrome p450, subfamily IIC, polypeptide 9 (CYP2C9), the two enzymes related to the metabolism of vitamin K and warfarin,7,15 account for genetic variabilities in the anticoagulation response.16-19 The inclusion of VKORC1 and CYP2C9 to a dosing algorithm may enable better prediction of warfarin dosing than when demographic data alone are used.20-22 Several warfarin dosage formulae and dose determination algorithms that include genetic information have recently been reported.22,23
The use of genetic polymorphisms in regulating warfarin dosase has recently been studied in Asian patients including Japanese,24-26 Chinese27,28 and Koreans.29-31 However, these previous algorithms have focused on the generalized warfarin dosage adjustment without considering tailored individual target INR. Accordingly, we examined the single nucleotide polymorphisms (SNPs) in VKORC1 and CYP2C9 in Korean stroke patients, and analyzed their effects on warfarin dosing to optimize stable INR. We additionally attempted to formulate a warfarin dosage equation that may be practically used for stroke patients.
Methods
Subjects
Between November 2009 and August 2010 at the Asan Medical Center neurology outpatient clinic, we prospectively enrolled patients who were diagnosed as having ischemic stroke confirmed by brain imaging studies, aged ≥18 years, and administered warfarin regularly under strict instructions for secondary prevention of ischemic stroke. Although INR level 2-3 has been considered 'optimal' for secondary prevention of stroke,32 considering its variation secondary to diet and occasional higher dosages needed in certain patients, such as those with mechanical valve replacement,32 we considered the INR level 1.7-3.5 'relatively stable',33 and enrolled patients who displayed this INR level on more than three consecutive measurements over at least 3 months before enrollment. We excluded patients who were 1) considered incompliant or anticipated to alter anticoagulation therapy (i.e., due to scheduled surgery); 2) severely dementic or aphasic so that reliable communication was not possible; 3) taking long-term medication that could potentially influence anticoagulation intensity, such as anti-epileptics, antibiotics, anti-inflammatory drugs, and alcohol; 4) diagnosed with underlying diseases potentially affecting the level of coagulation, such as vitamin K deficiency, malabsorption syndrome, malignancy; and 5) those who refused to consent. Patients with non-detectable R and S warfarin levels on the sampling date were additionally excluded. The study was approved by Ethics Committee of the Asan Medical Center, and written informed consent was obtained from all the subjects.
We reviewed medical records and interviewed individual patients. Data obtained were reasons for prescribing anticoagulation (e.g., arrhythmia, valvular heart disease), recent myocardiac infarction, heart failure, recurrent strokes despite antiplatelet use or severe intracranial stenosis, daily warfarin dose prescribed, and the warfarin medication schedule. Demographic features and risk factors for stroke were also recorded including age, gender, hypertension (defined as receiving medication for hypertension or blood pressure >140/90 mmHg with repeated measurements), diabetes mellitus (defined as receiving medication for diabetes mellitus, fasting blood sugar ≥126 mg/dL or post-prandial 2 hours≥200 mg/dL with two separate measurements), hypercholesterolemia (defined as receiving cholesterol-reducing agents, overnight fasting low density lipoprotein (LDL) level >100 mg/dL or overnight fasting cholesterol level >200 mg/dL), and current cigarette smoking (defined as smoking within six months of enrollment). The body mass index (BMI) was measured in each patient.
Blood sampling and determination of biological phenotype
Since 2000, we have provided patients receiving anticoagulation therapy official education with an instruction program. At the time of enrollment, a well-trained nurse confirmed that the patient had received the instructions. Blood samples (10 cc) were obtained 12 hours after warfarin intake. We measured INR using standard methods.34 To determine patient drug compliance, the plasma warfarin levels of patients (R- and S-warfarin) were measured using chiral-phase high-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS; ThermoFinnigan, San Jose, CA, USA), as described elsewhere, with minor modifications.35,36 While all patients followed our diet education program, serum vitamin K levels (phylloquinone) were additionally measured to adjust for the possible confounding effects of diet on INR, using LC-MS/MS with atmospheric pressure chemical ionization, as described elsewhere.37
We assumed three variables as biological phenotypes influenced by genotype: INR, daily maintenance warfarin dose, and warfarin response index (WRI). WRI was defined as INR divided by the daily maintenance warfarin dose (mg). Since WRI seems to be a more reliable variable reflecting individual response than INR or warfarin dose (see below), the index was employed as the biological phenotype in subsequent analyses. Based on WRI data obtained on the date of blood sampling, patients were divided into three groups: high (upper, 33.3%), intermediate (33.4-66.5%), and low (lower 33.3%) using two cutoff values. Novel SNPs were examined in two (high vs. low) groups.
Determination of genotype
We searched known SNPs related with warfarin metabolism from published papers,31,38 database (http://www.ncbi/nlm.nih.gov/). We also sequenced whole VKORC1 genes for confirmation of the presence of unreported SNP or linkage between the high WRI and low WRI groups. We selected 26 SNPs in CYP2C9 (three sites; 430C>T (rs1799853), 1075A>C (rs10 57910), 1076T>C), VKORC1 (14 sites; 381C>T, 861C>A (rs17880887), 2653G>C, 3673A>G, 5496G>T, 5808T>G (r17882154), 6009C>T, 6484T>C (rs9934438), 6853C>T (rs17886369), 7566T>C, 8767G>C, 8814T>C, 9041G>A (rs17880624), 9071G>T), gamma-glutamyl carboxylase (GGCX, nine sites; eight SNPs and one microsatellite; 268A>T, 412G>A, 6258A>G, 8015C>T, 8016G>A, 8240A>G, 8445C> T, 13875A>G, CAA repeat).
Sequencing method (resequencing analysis for the detection of genetic variations) was used to determine genotype. For sequencing analysis, genomic sequences were obtained from the GenBank (http://www.ncbi/nlm.nih.gov/) database and PCR primers were designed using the Primer3 software (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) to amplify coding regions and promoter regions (≥500 bp upstream from exon 1) of VKORC1 and CYP2C9 genes (PCR primer information used in this study is available on request). PCR amplification reactions were performed in 20-µL total volume containing 20 ng genomic DNA, 0.15 µM of each primer, 100 µM of each dNTP, 1 unit of AmpliTaq Gold™ (Applied Biosystems, Foster City, CA, USA) and reaction buffer with 1.5 mM MgCl2. PCR was performed at 94℃ for 5 minutes, followed by 35 cycles at 94℃ for 45 seconds, 55℃ for 30 seconds, 72℃ for 1 minute and final extension at 72℃ for 10 minutes. The PCR products (1 µL) were directly used as template for sequencing using BigDye™ terminator v2.0 cycle sequencing kit (Applied Biosystems) on an ABI3730 DNA sequencer according to manufacturer's instructions. DNA polymorphisms were identified using the PolyPhred program (http://droog.gs.washington.edu/PolyPhred.html) after sequence chromatograms were basecalled with the Phred program and assembled with Phrap. In the VKORC1, we searched unknown SNPs related with clinical phenotype. We selected high and low WRI groups and analyzed whole VKORC1 sequence to try and identify unknown SNP between groups. Also, we confirmed genotype using the Taq-Man SNP genotyping assay (Applied Biosystems) according to the manufacturer's instructions. Two polymorphic sites (rs7294 for VKORC1 gene and rs1057910 for CYP2C9) were selected and genotyped for reconfirmation. Fluorescence intensities were read on a 7900HT Sequence Detection System (Applied Biosystems), and genotypes were determined using SDS 2.1 software (Applied Biosystems).
Statistical analyses
Genotyping data were tested for deviation from the Hardy-Weinberg equilibrium using the Chi-square test. We analyzed log-transformed data of variables for biological phenotypes, specifically, INR, warfarin dose, and WRI, which were skewed. Fisher's exact test was used to compare the allelic frequencies of VKORC1 and CYP2C9 variants among patients with different variables. We calculated the effective sample size as 155.35 using calculation fomula39 (power 90%, α=0.05, correlation coefficients=0.1, Z β=1.282, Z α/2=1.96, Fisher's transformed Z=0.5*ln (1.1/0.9)=0.1003, sample size=([1.96+1.282]/0.1003)2 +3=155.35).
Relationships between items such as age, body weight, BMI, and WRI and warfarin, respectively, were analyzed using Pearson correlation while the relationship between WRI and warfarin dose with items such as gender, hypertension, diabetes, hypercholesterolemia, and cigarette smoking were tested with t-test. Multivariate linear regression analysis was performed to determine the formula for warfarin dosage. All statistical analyses were performed using SPSS 14.0 (SPSS, Chicago, IL), with the level of statistical significance being set at P<0.05.
Results
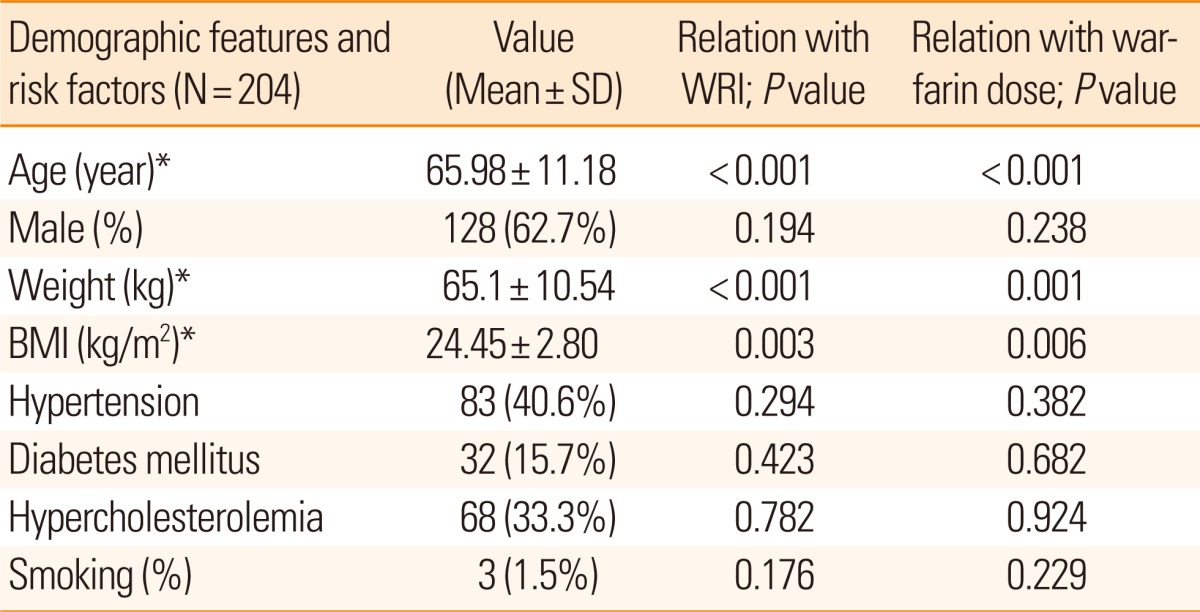
Our study included 204 patients with a mean age of 66 years (SD, ±11.2) ranging from 25 to 87 years, including 128 males (63%). The mean BMI was 24.5 kg/m2 (SD, ±2.8; range, 13.7 to 32.2 kg/m2). The reasons for anticoagulation included arrhythmia (150 patients, 73.4%), recurrent strokes despite antiplatelet use (39 patients, 19.2%), valvular replacement (10 patients, 5%), and others (five patients, 2.4%). All patients showed good drug compliance. The mean INR was 2.22 (SD, ±0.56; range, 1.12 to 3.74), mean daily maintenance dose of warfarin was 3.92 mg (SD, ±1.54; range, 1.29 to 10.0), and the mean WRI was 0.648 (SD, ±0.292; range, 0.178 to 1.806).
As shown in Table 1, WRI was closely related to old age (P< 0.001), BMI (P=0.003), and body weight (P<0.001). Mean R- and S-warfarin levels were 0.188 (SD, ±0.110) and 0.081 µg/mL (SD, ±0.059), respectively and they were significantly correlated to WRI (P<0.001, respectively), while the vitamin K level (mean, 0.830 g/mL; SD, +0.581) was not (P=0.123).
Demographic features and risk factors, and correlation with biological phenotype (warfarin dose and WRI)
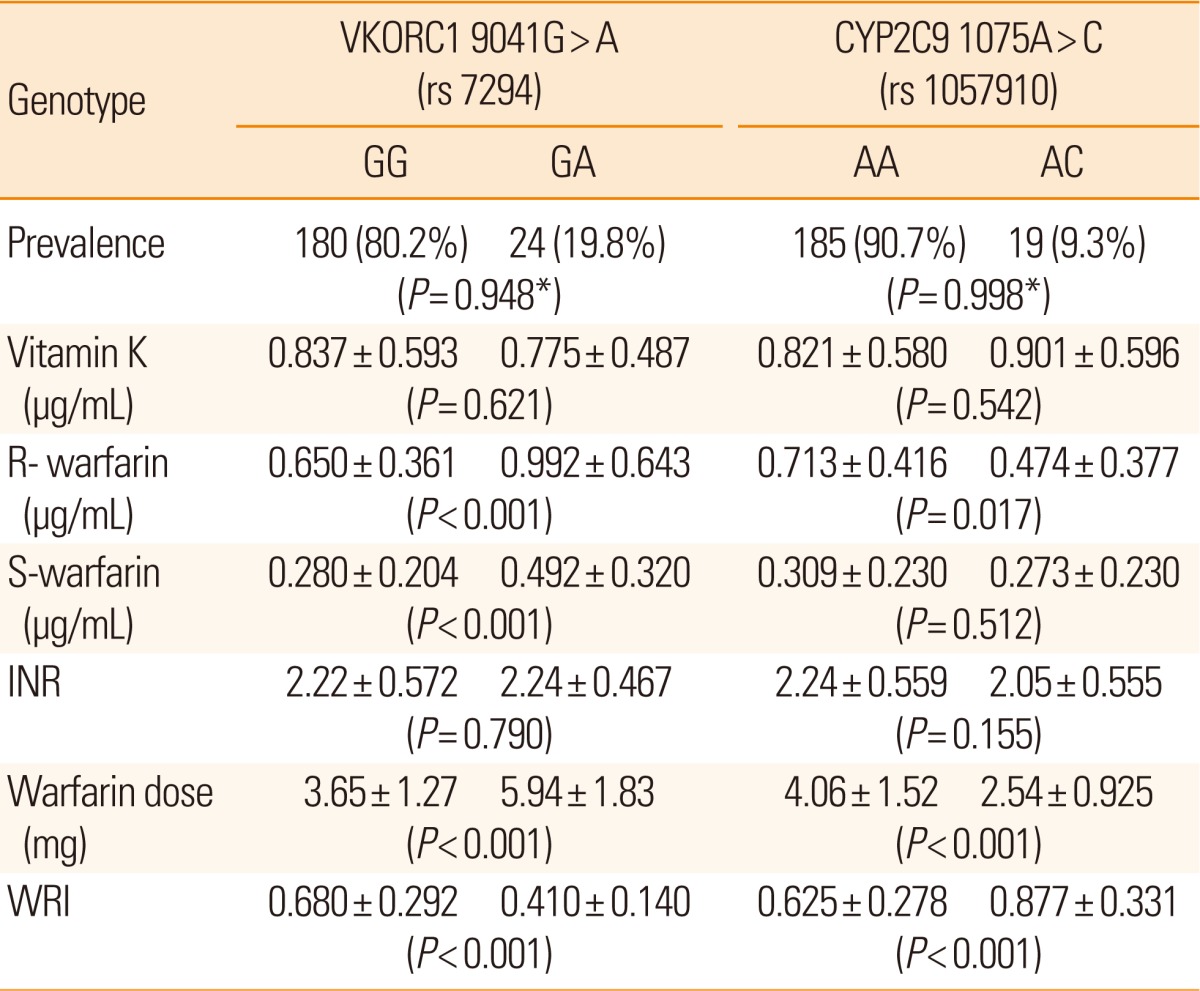
We analyzed the prevalence of all targeted genotypes (data not shown). Seven SNPs were related to a significant difference of WRI (6 SNPs located in VKORC1 (381T>C, 3673G>A, 6484T>C [rs9934438], 6853C>G [rs17886369]. 7566C>T, 9041G>A [rs17880624]) and one in CYP2C9 [1075A>C]). In terms of genotype prevalence, 24 patients (11.8%) were heterozygotic in six SNPs of VKORC1 and 185 patients (90.7%) were heterozygotic in one SNP of CYP2C9. The six SNPs in VKORC1 were 100% interlinked (r2=1). In view of the finding that 9041 G>A (rs17880624) displayed the most consistent results with repeated genotype tests using the TaqMan SNP genotyping assay (Table 2), we selected this site for subsequent analysis.
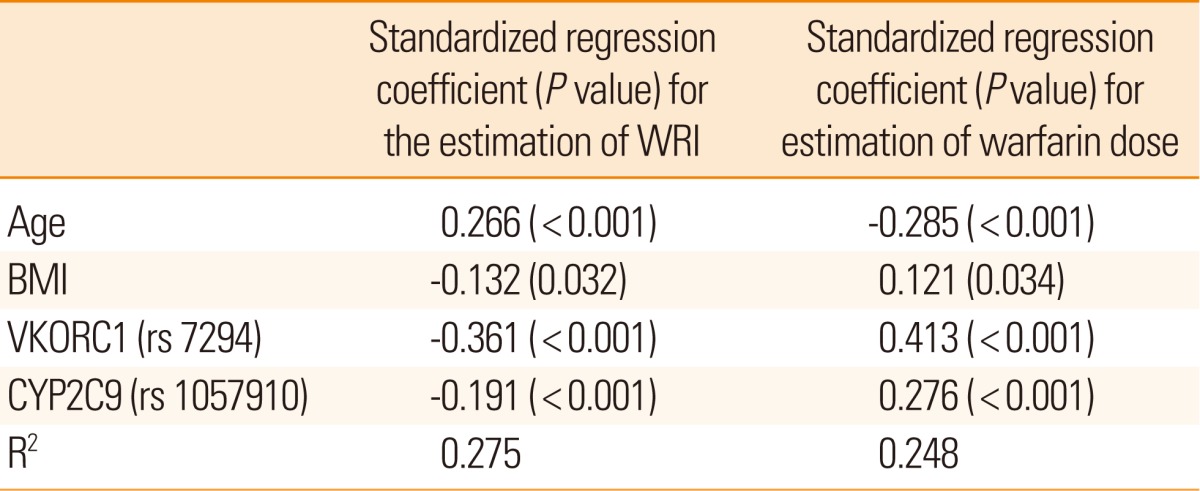
The high prevalence of heterozygotic VKORC1 9041 G>A (rs17880624) was closely associated with low WRI (P<0.001), and prevalence of heterozygote CYP2C9 1075A>C (rs1057910) with high WRI (P<0.001) (Figure 1, Table 2). Other SNPs did not show notable correlation with WRI or other biological phenotype variables examined. Moreover, in subgroup analysis, we sequenced the VKORC1 gene in patients with the high (upper 33.3%) and low (lower 33.3%) WRI, but failed to identify novel SNPs that were significantly associated with biological phenotype variables. Multivariate linear regression analysis identified four variables that independently influenced the WRI value: age, BMI, and two SNPs in VKORC1 and CYP2C9 (R2=0.275, multivariate linear regression, Table 3). Using regression coefficients obtained from the above results, we established the warfarin dosage formula, as shown in Figure 2A. We additionally performed the above analysis using warfarin dose instead of WRI as a phenotype variable (Figure 2B). The results obtained were similar.

WRI and warfarin doses in different genotypes. (A) WRI (left) and warfarin doses (right) in VKORC1 rs7294; homozygote VKORC1 rs7294 is associated with significantly higher WRI (P< 0.001) and lower warfarin dose (P< 0.001) than heterozygote. (B) WRI (left) and warfarin doses (right) in CYP2C9 rs1057910; homozygote CYP2C9 rs1057910 is correlated with significantly lower WRI (P< 0.001) and higher warfarin dose (P< 0.001) than heterozygote.
Contribution of age, BMI and selective genetic variations to biological phenotype using multivariate linear regression analysis
Discussion
We investigated the factors influencing WRI (and warfarin dose) in a relatively large number of homogeneous Korean stroke patients. Young age, high BMI, heterozygote VKORC1 (9041G>A, rs17880624), and/or homozygote CYP2C9 (1075A>C, rs1057910) contributed to low WRI, indicating that patients with these characteristics have low anticoagulation responses to a given warfarin dose. To minimize the confounding effect of non-compliance, we selected patients who had been formally educated with the institutional protocol. Furthermore, since diet preference may influence the INR level,40 we concomitantly measured the serum vitamin K level to adjust potential dietary effects on WRI. The vitamin K level did not influence the anticoagulation response. In addition, R- and S-warfarin levels were highly correlated with WRI (P<0.001), illustrating the patients' excellent drug compliance.
In this study, our aim was to determine the tailored intensity of anticoagulation response in Koreans and to provide a practical warfarin dosage equation using WRI. The information about SNPs agreed with a previous genetic association study in Asian population showing the importance of genetic variability and prevalence of selected SNPs (VKORC1 9041G>A and CYP2C9 1075A>C). However, the linkage in VKORC1 SNPs has not been discovered in the previous study in Korean population.31 In the GGCX gene, we could not find any significant SNP, which has shown inconsistent results in previous Japanese studies.24,26
Our data were also consistent with results from studies with a multi-ethnic population14,38,41 reporting that patients with at least one heterozygote in several SNPs in VKORC1 and CYP2C9 1075A>C require a high warfarin dose. While 9041G>A was not linked with other SNPs (at positions 381, 3673, 6484, 6853, and 7566) among European-American,38 our result in Koreans showed strong correlation (r2=1) among six SNPs in VKORC1. Also, we could not find significant influence of CYP2C9 (1076T>C or 430C>T) on warfarin dose although this position was associated with warfarin dose in the previous studies.38,41-43 These differences may be caused by ethnic genetic variability.
In contrast to previous studies employing warfarin dose only as a phenotype variable44-46 we used WRI, a variable that provides a comprehensive estimation of the warfarin dose in relation to target INR. While our main results are similar to those obtained using warfarin dose only, using WRI as a phenotype variable has several advantages. First, the predictability of the formula using WRI (R2=0.275) was higher than that with warfarin dose (R2=0.248). Second, WRI distinguishes inter-individual biological phenotypes more precisely. For instance, patients with INR 1.5 receiving 10 mg warfarin should be biologically different from those with INR 4 receiving the same dose. Third, the formula using WRI as a variable is more adaptive in daily clinical practice. Since this formula includes the INR level, the warfarin dose can be adjusted according to the clinical need to alter target INR, such as when new ischemic or hemorrhagic strokes develop. Therefore, our data are practically applicable to general Korean population to calculate stable warfarin dose.
Although our data may not be directly applicable to other ethnicities with different genetic background, our results may encourage further studies to develop the warfarin dosing formula in different ethnicities.
Notes
The authors have no financial conflicts of interest.
This study was supported by a grant from Brain Research Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology of Korea (M103KV010010 06K2201 01010).